
The task
Every biological system uses fundamental physics to perform its daily function. We study how mechanics is involved and sometimes even responsible for the correct and robust interaction between the different biological components. Although cell and tissue mechanics has been a research focus during the past two decades, we only start to scratch the surface of all the intriguingly complex dependencies between biochemical signaling, mechanical forces and viscoelastic properties. The quantitative description and the mathematical modelling of the complex interaction inside cells and tissue are the focus of the lab. Therefore we develop new measurement methods to quantify forces and tension in 3D tissue, membranes and filaments. Furthermore, we investigate new microscopy and analysis methods and integrate these with quantitative measurements of the viscoelastic properties in cells can tissue.
A main aim is to decipher the fundamental physical processes leading to stability and robustness despite the complexity and highly non-equilibrium and non-linear nature of any living system. We hope to discover new physics in observing and describing the complexity of the living world.
Every biological system uses fundamental physics to perform its daily function. We study how mechanics is involved and sometimes even responsible for the correct and robust interaction between the different biological components. Although cell and tissue mechanics has been a research focus during the past two decades, we only start to scratch the surface of all the intriguingly complex dependencies between biochemical signaling, mechanical forces and viscoelastic properties. The quantitative description and the mathematical modelling of the complex interaction inside cells and tissue are the focus of the lab. Therefore we develop new measurement methods to quantify forces and tension in 3D tissue, membranes and filaments. Furthermore, we investigate new microscopy and analysis methods and integrate these with quantitative measurements of the viscoelastic properties in cells can tissue.
A main aim is to decipher the fundamental physical processes leading to stability and robustness despite the complexity and highly non-equilibrium and non-linear nature of any living system. We hope to discover new physics in observing and describing the complexity of the living world.
The current projects of the lab
Intracellular passive and active microrheology
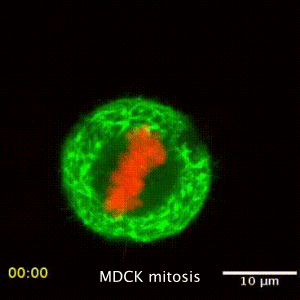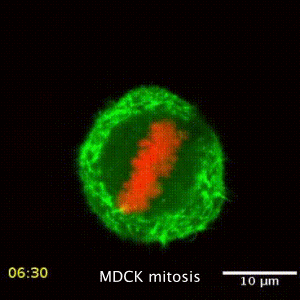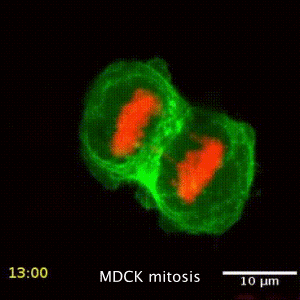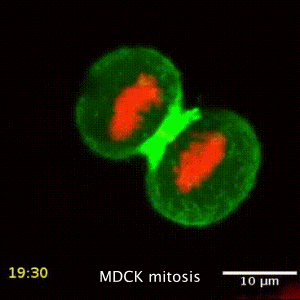
We are interested in the underlying mechanism of organelle distribution during cell division, migration and polarity, and try to understand these by studying the intracellular forces, the local changes of viscoelastic material properties and the active, non-equilibrium processes driving the active and passive transport inside cells. While active microrheology allows direct access to the viscoelastic material properties, its combination with the passive motion analysis of the particle or organelle motion gives direct information about the processes driving transport. The rapid motion is driven by Brownian, hence thermal motion, while the slow processes are driven by active metabolic forces. The timescale separating these regimes depends on the cell type and situation studied. This led to the development of a mechanial fingerprint that allows to differentiate between celltypes.
We furthermore develop new methods to obtain both activity and passive material properteis from the spontanouse fluctuations of intracellular particles. The new quantity is coined Mean Back Relaxation (MBR), which allows to determine broken detailed ballance very easily.
We furthermore develop new methods to obtain both activity and passive material properteis from the spontanouse fluctuations of intracellular particles. The new quantity is coined Mean Back Relaxation (MBR), which allows to determine broken detailed ballance very easily.
Collective cell migration in development
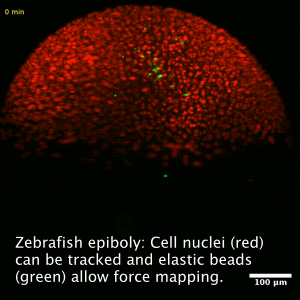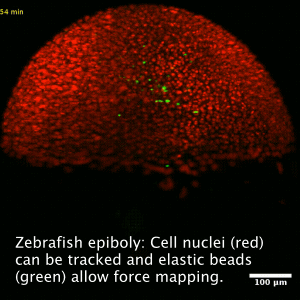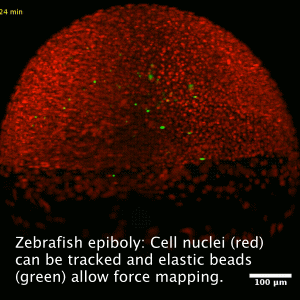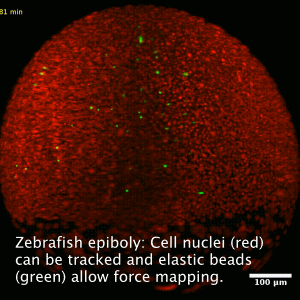
Collective cell migration is a fundamental process during embryogenesis and its initial occurrence in the zebrafish embryo, called epiboly, is an excellent in vivo model to study the physical processes involved in the collective cell movements which are important for the understanding of organ formation, cancer invasion, and wound healing.
In zebrafish, epiboly starts when the cells of the early embryo start actively spreading in a continuous movement toward the opposite pole of the spherical yolk-blastula system they fully cover the yolk.
In zebrafish, epiboly starts when the cells of the early embryo start actively spreading in a continuous movement toward the opposite pole of the spherical yolk-blastula system they fully cover the yolk.

We want to understand how this collective movement is organized from a physical point of view. What are the force generating mechanisms? Can we reconstruct the collective behavior applying rather simple physical models? In order to answer these questions, we use advanced imaging techniques as e.g. light sheet microscopy, analyze the data by tracking cells in the acquired images and perform viscoelastic measurements using optical tweezers, as well as recently developed elastic force sensors, that can be analyzed easily with out software BeadBuddy.
Collective cancer cell migration

Collective migration is a gold standard behavior that has been commonly observed during morphogenesis and during wound healing. Cancer cells move collectively in order to migrate and overcome the space constriction due to densely growing cells at the center. This is characterized by cell-cell adhesion differences, the extracellular matrix or cell-substrate interactions, and the intercellular cross-talks in-order to metastasize. The mechanisms and the physics behind the coordinated behavior of cells have been studied recently at the 2D level, and in-vivo tissue homeostasis. However due to the challenges involved in reproducing and imaging tumor spheroids; collective motion of 3D cancer invasion is not yet completely understood. With the use of Light Sheet Microscopy, we have been able to consistently image HeLa spheroids for long hours (~48h) at sub-cellular resolution. This has enabled us to look deeper into how the velocity of cells correlate with one another, via particle tracking over a period of time, as they radially arrange themselves in collagen.
“Forcing” changes in the adult stem cell transcriptome
Skeletal muscle is the most widespread tissue in the human body and supports essential functions e.g. breathing, swallowing and movement. Upon injury, muscle stem cells, also known as satellite cells (SCs), are activated and eventually regenerate muscle tissue by fusion of differentiated myogenic progenitor cells to form new myofibers. Disorders during this myogenic program may lead to severe muscular dystrophies. Whereas many signal transduction pathways are known to be responsible for SC fate decision, especially mechanical niche cues are far less understood. Therefore, we are tremendously interested in a variety of forces, which are exerted on SCs in muscle tissue and are responsible to wake SCs up from their quiescent state to exhibit all their great regenerative potential.
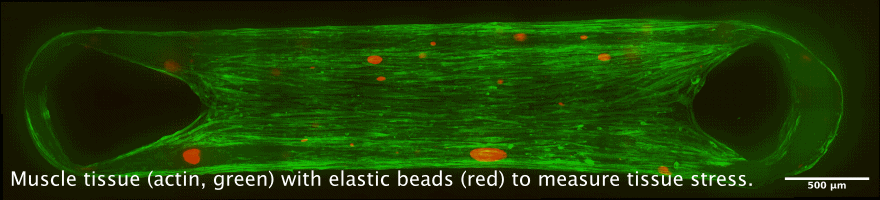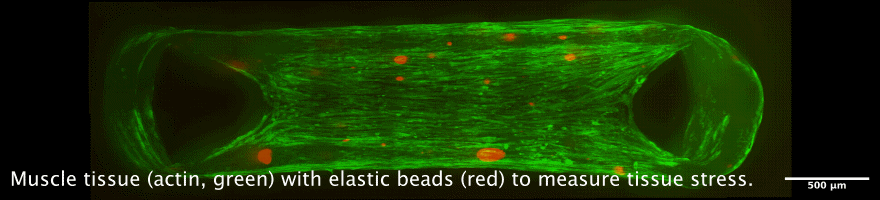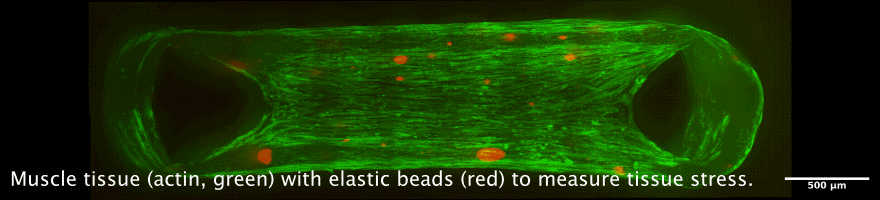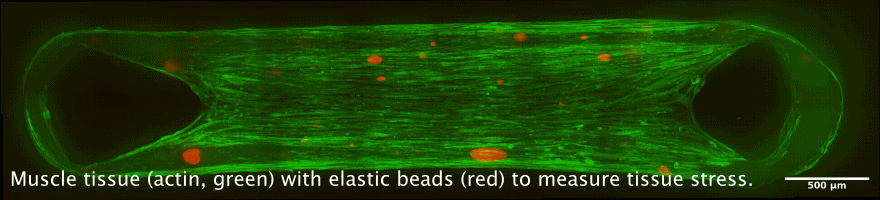

Tissue chambers for muscle and connective tissue
 The newly developped chamber allow to cultivate 3D tissue such as skeletal or heart muscle tissue in close proximity to a glass coverslip. This allows high resolution microscopy while maintaining a perfectly organized and living tissue. Interesting applications are the measurement of force generation in combination with cellular and nuclear mobility, for example during muscle development, or during heart diseases.
The newly developped chamber allow to cultivate 3D tissue such as skeletal or heart muscle tissue in close proximity to a glass coverslip. This allows high resolution microscopy while maintaining a perfectly organized and living tissue. Interesting applications are the measurement of force generation in combination with cellular and nuclear mobility, for example during muscle development, or during heart diseases.

A further possibility is the detection of cancer cell migration in structured 3D environments that resemble the connective tissue found in teh body. Here we coculture fibroblasts with cancer cells to study the invasion of these cells in the 3D tissue. In the video on the right you see marked nuclei of cancer cell migrating in the structures 3D tissue generated by fibroblasts. Some cells have an additional marking of actin in green. Cell divide and form metastasis during the 90h video.
 The newly developped chamber allow to cultivate 3D tissue such as skeletal or heart muscle tissue in close proximity to a glass coverslip. This allows high resolution microscopy while maintaining a perfectly organized and living tissue. Interesting applications are the measurement of force generation in combination with cellular and nuclear mobility, for example during muscle development, or during heart diseases.
The newly developped chamber allow to cultivate 3D tissue such as skeletal or heart muscle tissue in close proximity to a glass coverslip. This allows high resolution microscopy while maintaining a perfectly organized and living tissue. Interesting applications are the measurement of force generation in combination with cellular and nuclear mobility, for example during muscle development, or during heart diseases.
A further possibility is the detection of cancer cell migration in structured 3D environments that resemble the connective tissue found in teh body. Here we coculture fibroblasts with cancer cells to study the invasion of these cells in the 3D tissue. In the video on the right you see marked nuclei of cancer cell migrating in the structures 3D tissue generated by fibroblasts. Some cells have an additional marking of actin in green. Cell divide and form metastasis during the 90h video.
Instrument design and software development

A large part of our projects requires to develop new instruments, microscopes and combinations of optics and advanced biophysics tools. Often, we extend existing methods and commercial microscopes to yield new and better performance. Key elements here are optical tweezers and high precision position detection methods. Besides such instrumentation, additional software and analysis tools are developed. Here we have a strong focus on measurement and automation tools, as well as methods to infer forces and tension from deformed objects (Traction force microscopy, Bead deformation assays).
Special Thanks to all the Funding!







